Differential Scanning Calorimetry (DSC)
Differential Scanning Calorimetry (DSC) measures the heat capacity, Cp, of a molecule (the heat required to warm it, per degree) as a function of temperature. It is applied in the biotechnology industry to:
-
study the thermal stability of a wide array of protein or peptide therapeutics and vaccines
-
demonstrate comparability of higher order structure (HOS) for material from different manufacturing processes or manufacturing sites.
Specifically, DSC measures the difference in heat flow between a sample and reference chamber, as both chambers are slowly heated (or cooled). Both chambers are enclosed by an adiabatic insulator. The sample chamber contains the molecule of interest in an appropriate buffer, while the reference chamber contains the identical buffer. When the sample cell is loaded with a protein, slightly more heat will flow into the sample cell than the reference cell, even at temperatures far from the thermal unfolding ("melting") temperature, Tm. This occurs because energy flows into the protein to break the non-covalent bonds associated with its solvation and its secondary and tertiary structure. When the temperature reaches a point where the protein starts to unfold, the difference in heat flow grows substantially, reaches a maximum at Tm, and then decreases as unfolding completes, producing a peak in the heat flow vs. temperature plot (thermogram). The graph below shows some example data we collected on our Microcal VP-DSC instrument for lysozyme, a protein often used as a control to test instrument performance because it gives nicely reversible unfolding. Note that we also now have an automated VP-capillary instrument with similar sensitivity.

The high sensitivity of this instrument allows collection of data of this quality at a concentration of only 0.2 mg/mL even for a small ~14 kDa protein like lysozyme (the heat of unfolding is generally larger for larger proteins, so they typically give even better signal/noise ratios). The red curve is a fit of these data to a simple two-state unfolding model (all-or-none folding, without intermediates). That fit gives a Tm value (the mid-point of the unfolding transition) of 63.9 °C, a heat of unfolding ΔH0 = 104 kcal/mol, and a heat capacity change ΔCp = 220 cal/mole/deg (the difference in heat capacity between the unfolded and folded state).
The graph below shows the excellent agreement between repeat measurements (different fillings from the same stock), even at only 0.2 mg/mL.

Reversibility of unfolding
For proper interpretation of DSC data it is critical to know whether the unfolding is reversible or irreversible. The most common cause of irreversibility is irreversible aggregation of the unfolded molecules, but chemical degradation at elevated temperatures could also occur. Reversibility can be assessed experimentally by cooling the unfolded sample in the calorimeter and then scanning it again. Alternatively, visual examination of the sample removed from the cell after heating may show that it is obviously irreversibly aggregated or precipitated.
Consequences of aggregation after unfolding
It is fairly common for the protein molecules to aggregate, and even precipitate, after they unfold, especially at high sample concentrations. The aggregation/precipitation reactions will also be detected by the calorimeter, and these reactions usually release heat (a negative signal). When aggregation follows unfolding it is not possible to obtain a correct post-unfolding baseline and thus impossible to accurately measure the enthalpy of unfolding.
In addition, since aggregation may start before the unfolding is complete, it can cause a shift of the apparent Tm to lower temperatures. That is, the combination of a positive signal from unfolding and the negative signal from aggregation causes the maximum positive signal to occur at a temperature lower than the true thermodynamic Tm. The magnitude of the shift in Tm will depend on the protein concentration and also the scan rate. Note that for any given protein whether aggregation occurs after unfolding usually also depends strongly on the composition and pH of the formulation buffer.
As an example, the graph below overlays thermograms for a monoclonal antibody at four concentrations from 0.055 to 1.1 mg/mL from our non-automated VP-DSC instrument ("lollipop" cells), after normalizing for the protein concentration and aligning the graphs at 50 °C. At the higher protein concentrations, after the main unfolding transition at ~72 °C the heat capacity drops sharply and goes negative due to aggregation and precipitation. The position of the top of the positive peak is also shifted slightly to lower temperatures at the higher concentrations (but note that for other proteins the shifts can be much larger). These effects are sometimes called the "aggregation artifact". As shown in the inset, only at a very low concentration of 0.055 mg/mL do we see a post-transition baseline region, but the excellent sensitivity of our instrument allows good quality data to be obtained at that low concentration.
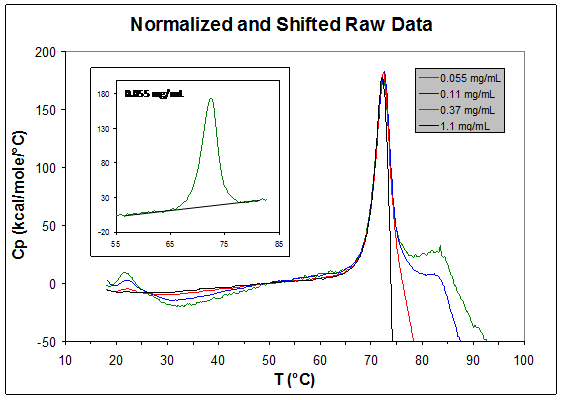
Clearly it is important to know whether such aggregation effects are occurring or the DSC data might be misinterpreted. Our automated VP-capillary instrument is much less sensitive to the post-melting aggregation events, but that lack of sensitivity is somewhat dangerous because it may lead to failing to realize that post-unfolding aggregation does occur.
DSC of monoclonal antibodies
DSC data for mAbs is particularly interesting because it sometimes resolves the unfolding of individual domains within the mAb structure, producing up to three peaks in the thermogram (from unfolding of the Fab, CH2, and CH3 domains). Typically unfolding of the Fab domain produces the strongest peak. The DSC profiles and relative stability of the Fc portion show characteristic differences for the human IgG1, IgG2, IgG3, and IgG4 subclasses [1].
The figure below shows some published data [2] for a mAb that gives 2 peaks at neutral pH (curve c) at 67 °C and 78 °C, and where we see major changes in the unfolding profile and shifts of the different transitions which are related to conformational changes triggered by the low pH used to elute it after binding to protein A (pH 2.7 for curve a, 3.5 for curve b).

For some mAbs and certain formulation conditions the thermograms may show only a single peak. However that doesn't necessarily mean that the entire antibody molecule is unfolding in a simple one-step process. The data shown below appear to show a single transition, but attempting to fit that single peak as a simple two-state unfolding transition gives a horrible fit.

A more detailed analysis shows this experiment can be well fitted as two successive two-state transitions for two independent folding domains with slightly different unfolding temperatures, as shown below.

DSC-related downloads
"Calorimetry study of a mAb that precipitates upon thermal denaturation", poster, WCBP, 2014.
[references]
[1] Garber, E. and Demarest, S. J. (2007). A broad range of Fab stabilities within a host of therapeutic IgGs. Biochem. Biophys. Res. Commun. 355, 751-757.
[2] Ejima, D., Tsumoto, K., Fukada, H., Yumioka, R., Nagase, K., Arakawa, T., and Philo, J. S. (2007). Effects of acid exposure on the conformation, stability, and aggregation of monoclonal antibodies. Proteins 66, 954-962.
